Diverse polypeptide sequences constitute an expansive reservoir with the potential for drug discovery. Antimicrobial peptides (AMPs) emerge as a promising avenue for combatting drug-resistant bacterial infections. Despite this potential, current advanced experimental technologies limit AMP screening to libraries of approximately 800,000 sequences. How to establish an efficient and precise methodology for the effective recognition of antimicrobial peptides within comprehensive short peptide libraries?
About the Author

Dr. Junjie Huang
Postdoctoral Fellow of International Research Center for Functional Polymers, International Campus, Zhejiang University
He has long been engaged in the research and development of machine learning accelerated materials and high-throughput experimental accelerated materials. He has published 7 articles in journals like Nat. Biomed. Eng., ACS Applied Materials & Interfaces and authorized 3 patents. In 2022, he won Hangzhou Qingshan Lake Material Genetic Engineering Young Scientist Award, Outstanding Academic Achievement of Zhejiang University International Campus.
This study introduces a novel machine learning pipeline, Sequential Model Ensemble Pipeline (SMEP), encompassing empirical selection, classification, ranking, and regression tasks for the identification of potent antimicrobial peptides within a hexapeptide-to-nonapeptide sequence library, spanning 500 billion samples over 27 days. The pipeline identified three leading antimicrobial hexapeptides, demonstrating significant bactericidal efficacy against a diverse range of clinically isolated drug-resistant bacteria. These peptides exhibited low off-target toxicity and displayed resistance-resistant properties. Furthermore, their therapeutic efficacy was substantiated in a mouse model of bacterial pneumonia. The findings underscore the effectiveness of the SMEP in systematically mining antimicrobial peptides with desirable properties from vast and complex peptide libraries.
01 Overview of SMEP
This study introduces a rapid antimicrobial peptide (AMP) recognition method using the Sequential Model Ensemble Pipeline (SMEP). SMEP integrates empirical judgment, classification, ranking, regression models, incremental learning model modification, and wet experiments (Figure 1). Six model peptides are employed for the recognition task from a vast library of 64 million peptides. Prior to identification, classification, ranking, and regression models are trained using literature data.
In the recognition phase, a refined 3.93 million polypeptide sublibrary is generated through empirical judgment (positivity; amphiphilic). The classification model categorizes peptides, and the 560,000 model predicts antibacterial activity. Subsequently, the sorting model pre-sorts antimicrobial peptides, with the top 500 selected for the final AMP library. The regression model predicts the Minimum Inhibitory Concentration (MIC) of these peptides, and the 10 peptides with the lowest MIC predictions (CRRI antimicrobial peptides) undergo wet testing.
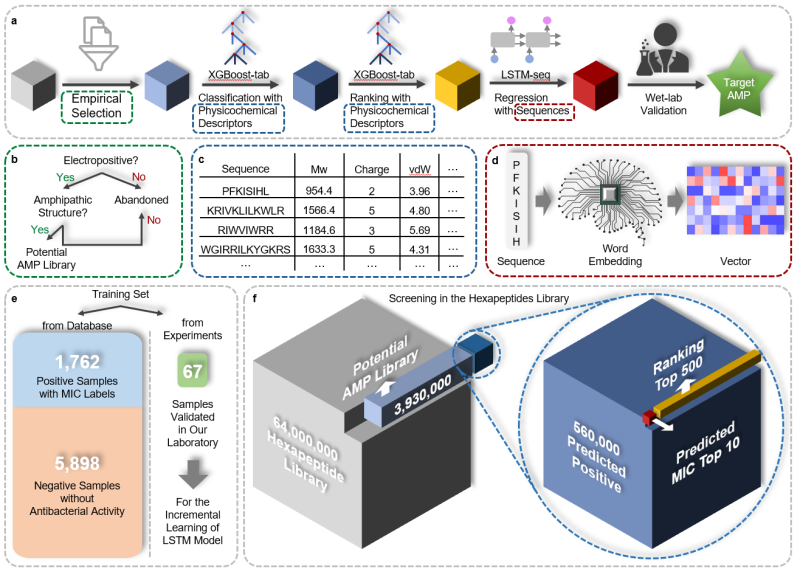
Figure 1. Overview of SMEP
02 Model Selection and Ablation Study
A dataset of 7,660 entries is employed to train/test classification, sorting, and regression models. XGBoost, XGBoost, and LSTM are chosen for classification, ranking, and regression, respectively. An ablation experiment validates the necessity of each module in SMEP (Figure 2B), reinforcing the pipeline's design rationale.
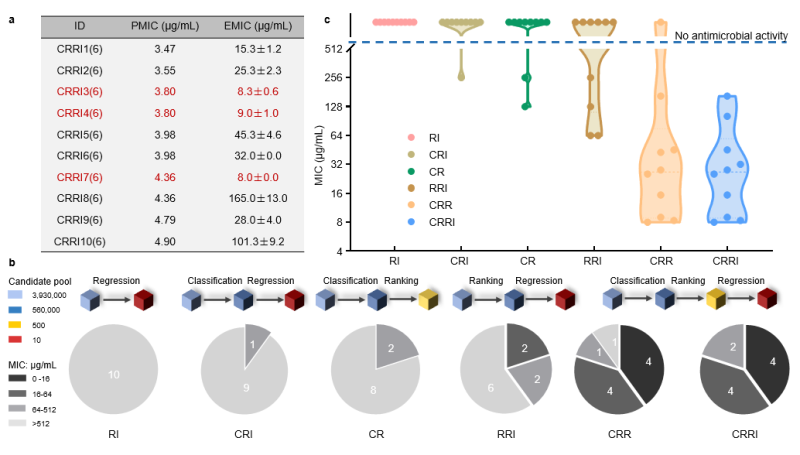
Figure 2: Ablation study
03 SMEP Generalization and In Vitro Testing of Antimicrobial Peptides
Without modification, SMEP efficiently recognizes heptapeptide, octapeptide, and nonapeptide AMPs in just 19 days, achieving a 98.2% success rate. Testing the most promising hexapeptides reveals robust antibacterial properties against multidrug-resistant strains with low toxicity and resistance induction (Figure 3A, Figure 4).
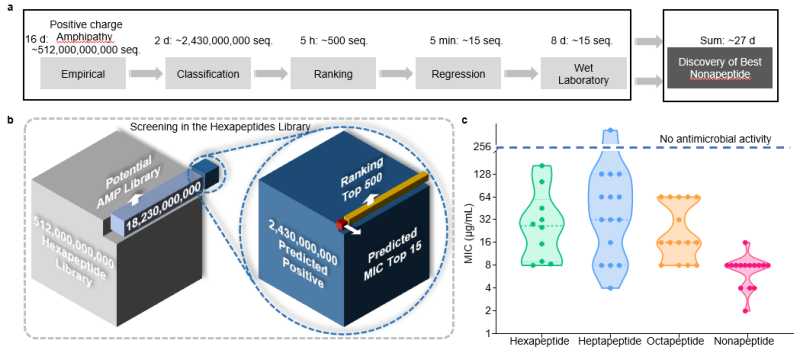
Figure 3. SMEP generalized heptapeptide, octapeptide, and nonapeptide antimicrobial peptide
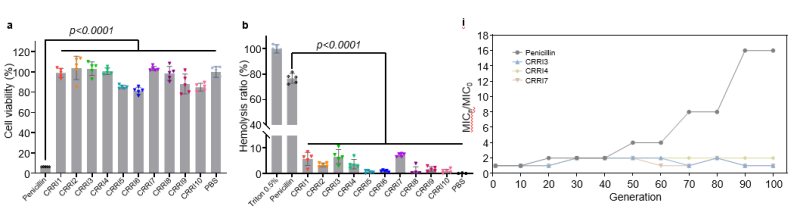
Figure 4. In vitro test of antibacterial hexapeptide
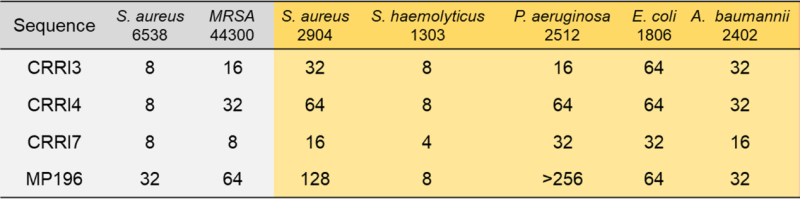
Table 1. MIC of AMPs against various strains
04 In Vivo Treatment Test of Antimicrobial Peptides
In acute pneumonia models, atomized administration of antimicrobial peptides (CRRI3, 4, 7 groups) results in a 99.8% reduction in lung bacteria and improved lung health. Chronic pneumonia models exhibit a 98.1% reduction in pulmonary bacteria, showcasing the excellent in vivo therapeutic efficacy of antimicrobial peptides.
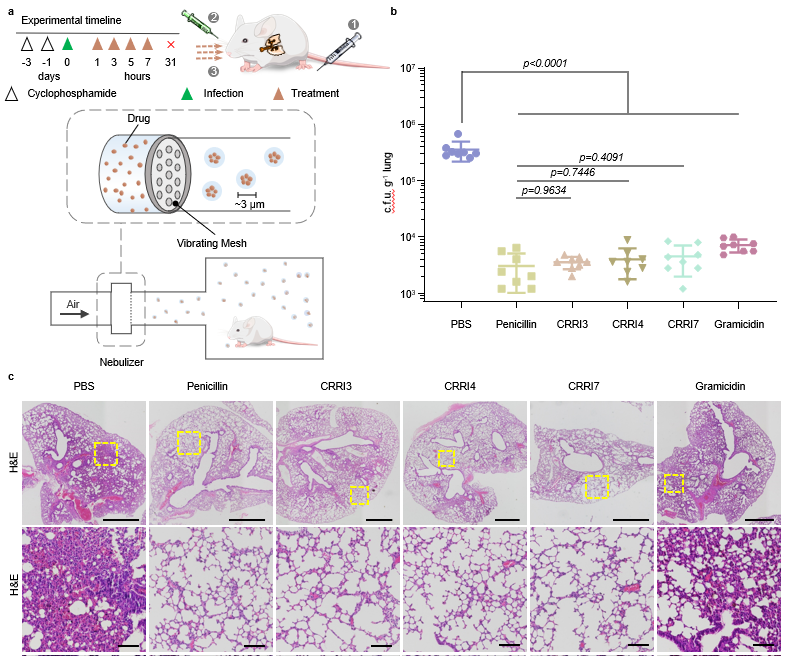
Figure 5. In vivo treatment test of antimicrobial peptides.
Author's Note
Embarking on the intersection of biomedical engineering and computer science marks my pioneering venture into uncharted territory. This multidisciplinary endeavor presents inherent challenges, notably the pronounced domain differences between computer science and biomedical engineering. The initial stages were characterized by significant communication barriers due to the unfamiliarity of computer scientists with biomedical research and my limited knowledge in computer science. Overcoming these disparities required tenacity, with both teams diligently acquiring insights into each other's domains, fostering a shared understanding of ideas.Beyond communication hurdles, groundbreaking initiatives inherently risk becoming entangled in complexity, often necessitating a reevaluation or even a complete restart. Enduring a year of repeated setbacks and realizing that incremental adjustments fell short of our research objectives, our team boldly dismantled the existing design paradigms. We meticulously reconstructed the model architecture, resulting in the realization of our scientific goals.
Embarking on uncharted territories demands courage, particularly when pursuing innovation. This journey, albeit challenging, holds the promise of a direct route to unparalleled heights.
Academic Achievement Award
International Campus selects the most commendable academic achievements of the year by the end of each year. In 2022, a total of 10 research achievements including this series of research were selected.

Article and Photograph: HUANG Junjie;
Editing: REN Yue, LI Zhenming, LI Yinan;
Editing in charge: XUE Qian;
Reviewer: WU Fengbin





